Before we get into the meat of the paper, a bit of an introduction is called for. The single, most basic lesson that is taught about the way that genes work in the cell is known as the Central Dogma: DNA is transcribed to produce RNA, and RNA is translated to produce protein. It's a nice, clearly stated concept, and it has the added virtue of often being true. DNA is transcribed into RNA, and the RNA is "read" in the cell's ribosomes to produce the various proteins that carry out most of the cell's functions. The Central Dogma is simple and relatively easy to understand.
Like most statements that are simple and easy to understand, the Central Dogma is not entirely true. There are some cases where the RNA is the final product, and does something else besides code for a protein. The RNA sometimes gets edited before it is used to make a protein. Some of the DNA has functions other than directly making protein. Genes work in more complex ways than those specified in the Dogma, and the Dogma doesn't cover all of the important parts of what goes on in the cell.
One of the things not covered by the Central Dogma is the matter of gene regulation. This is an important process in all living things, and is particularly important in complex multicellular organisms. Every cell has all of the DNA needed to do everything that the organism does, but not every cell actually does everything. In fact, after the early stages of development, no cell does everything. Different types of cells develop in different tissues and organs, and need different amounts of different proteins at different times.
I'm not going to get into the fine details of gene regulation here. For the purposes of understanding this one paper, it is enough to know that different cell types are going to need to make different amounts of various proteins at various times. Since these proteins are made from RNA, we can find out active a given gene is in a cell by looking to see how many copies of the RNA transcript for that gene are floating around in the cell.
Microarray analysis provides us with a method for comparing the amount of DNA expression for various genes in the same types of cell in two different species. The details are fairly complex, and you don't really need to understand them to get the basic gist of what was done in this paper, but if you want to know more, you can look here.
In the experiment reported in the Science article, the researchers used microarrays to examine the relative expression of different genes in humans and chimps in a number of different tissues - heart, kidney, liver, testis, and brain.
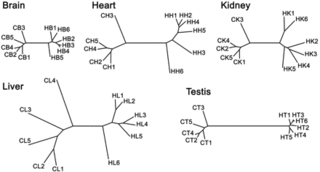
On the left, I have reproduced figure 1 from the Science article. This figure shows the differences in gene expression observed in the various individuals sampled. The samples that begin with "C" are chimps, and those that begin with "H" are human. The lines indicate the size of the difference, with short lines indicating little difference, and long lines indicating a great deal of difference. These results are quite interesting.
One of the things that they show is that in the sampled tissues, the greatest similarity in gene expression levels between humans and chimps is actually in the brain, while the greatest differences are in the liver. This probably isn't what you might have expected to see. After all, the differences in thought and behavior are generally considered to be enormous between humans and chimps. So what do these results actually indicate.
In the opinion of the paper's authors, these results demonstrate the importance of neutral changes in evolution. In such a model, most genetic change accumulates in places where it does not positively or negatively affect survival. The heart, the kidneys, and the liver function more or less the same in humans and chimps, and the figure shows that the differences in gene expression within humans and within chimps are similar to the difference between the two groups. Such a result indicates that selection has not been a large factor driving the divergence in expression in those tissues. Selection does appear to have been a factor in the differences in the testis, and it may have been in the brain.
So what might all this mean? The authors of the paper sum it up well:
we find that the patterns of evolutionary change in gene expression are largely compatible with a neutral model, in which different levels of constraints acting in different tissues add up for single genes. These evolutionary constraints act in a similar manner on the coding regions of DNA sequences and thus lead to parallel patterns in expression and sequence evolution. In contrast to the overall picture of selective neutrality, two examples of putative positive selection stand out. First, testis shows an excess of expression differences between species and an enrichment of both expression and amino acid sequence differences on the X chromosome. Second, the brain, although under more constraints than the other tissues, has an excess of gene expression and amino acid changes on the human lineage compared to other tissues. This suggests that evolutionary changes at both the level of gene regulation and the level of protein sequence have played crucial roles in the evolution of certain organ systems, such as those involved in cognition or male reproduction. Consequently, the modest number of sequence differences in genes between humans and chimpanzees cannot be taken as evidence that regulatory changes would necessarily be more important than structural protein changes during human evolution. Rather, both types of changes are likely to have acted in concert.
It's going to be interesting to see what the response to this paper is. In some ways, the findings back up some of the things that we've long suspected about the divergence of chimps and humans - in particular, that the differences might be mainly due to selection acting on a fairly small number of traits. In other ways, such as the suggestion that changes in proteins might be as important as changes in gene regulation, this paper may challenge the traditional model of human/chimp divergence. This kind of mixed result is kind of nice to see. It hints that we haven't been entirely wrong in the past, but that there is still plenty more to learn about what drove the speciation process that split us from the chimps.
Reference:
Parallel Patterns of Evolution in the Genomes and Transcriptomes of Humans and Chimpanzees
Philipp Khaitovich, Ines Hellmann, Wolfgang Enard, Katja Nowick, Marcus Leinweber, Henriette Franz, Gunter Weiss, Michael Lachmann, and Svante Pääbo
Science 16 September 2005: 1850-1854.

1 comment:
>> ...in particular, that the differences might be mainly due to selection acting on a fairly small number of traits.<<
If only a relatively few genetic changes differentiated proto-human from ur-chimp back at the start of the Great Schism, does that imply that these mutations were not that vastly improbable? If so, then another crucial question arises: what were the environmental differences that favored these changes _this time_?
Post a Comment